Research Themes
1. Evolution of Extreme Longevity in Eusocial Species
We investigate how reproductive castes in insects and mammals evolve extraordinary lifespans despite high reproductive output, minimal senescence, and minimal ageing phenotypes. Using genomics, transcriptomics, epigenomics, and mobilomics, we have identified molecular pathways and regulatory strategies that enable this trait to evolve multiple times.
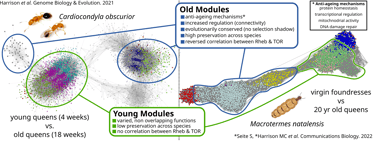
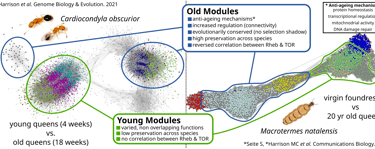
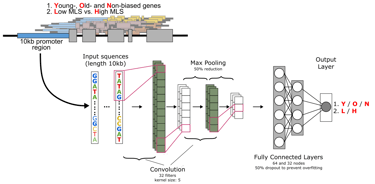
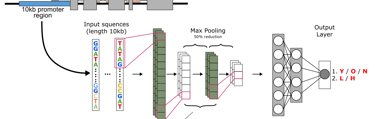
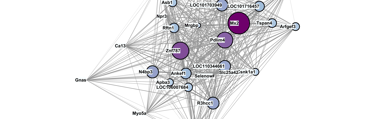
Fig. 1 Convergent co-expression network structures in long-lived ant and termite queens
Predicting lifespan from promoter sequence features with a convolutional neural network (CNN).
Question
Methods & Data
Preliminary Findings
On-going & Preliminary Work
Can promoter sequence features predict age-biased expression in mammalian tissues and maximum lifespan?
Convolutional neural networks trained on promoter sequences from 20 rodent species, plus liver age-expression datasets.
Initial classifier distinguishes high vs low lifespan species with ~73% accuracy; age-biased expression classification in naked mole-rat liver ~84%.
Expanding model to more species; implementing more sophisticated models (e.g. transformers); exploring regulatory motifs for intervention in humans.
Next Steps
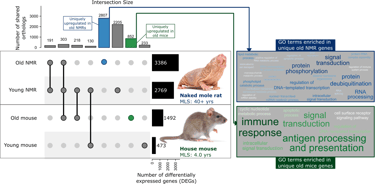

Do age-related gene expression patterns in long-lived mammals differ from those in shorter-lived mammals?
RNA-seq datasets from young vs old individuals of long-lived naked mole-rats (NMR) and mice; differential expression and network analyses.
2807 genes exclusively upregulated in old NMRs were enriched for transcriptional regulation and proteostasis. A module of the NMR gene network contained hub genes with roles in disease (Mx2, Pdlim4) and transcriptional regulation
Further sampling; network comparisons; integration with epigenetic data.

2. Comparative Genomics and Ageing in Mammals
Our current work focuses on identifying and characterising genomic and regulatory features that predict lifespan and ageing trajectories in mammals. We are particularly interested in long-lived species such as the naked mole-rat, certain bat and rodent species, and other mammals that defy typical ageing patterns. We are using machine learning models, promoter sequence analysis, and gene expression data to explore the evolutionary basis of longevity and to predict maximum lifespan from genomic data.
3. Towards Translational Ageing Biology and Health
By linking molecular signatures of long life from non-eusocial mammals to human ageing, we aim to uncover biomarkers and regulatory features of ageing pathologies. This translational angle may ultimately inform interventions for age-related diseases and contribute to strategies for healthy ageing in humans.
